Scientific Workflow Use – Towards Open Methods, by Richard Littauer
Maria Neicu - September 27, 2011 in Guest Post, Tools
 This summer I did an internship with DataONE, or the Data Observation Network for Earth, which is a US NSF-funded initiative aimed at “ensuring the preservation and access to multi-scale, multi-discipline, and multi-national science data.”
This summer I did an internship with DataONE, or the Data Observation Network for Earth, which is a US NSF-funded initiative aimed at “ensuring the preservation and access to multi-scale, multi-discipline, and multi-national science data.”
It is a virtual organization dedicated to providing open, persistent, robust, and secure access to biodiversity and environmental data. I was one of eight interns working on various projects – my project involved studying scientific workflows. Specifically, I was tasked with understanding how they could be categorised and how they are being used by the scientific community. I recorded my internship work on my Open Notebook, and presented (slides) on this work in progress at the Open Knowledge Foundation Conference in Berlin.
What are Workflows?
I had a Linguistics background before undergoing this project, and understandably I spent a long while going through the relevant literature trying to understand what exactly scientific workflows are. For those of you who don’t know (like I didn’t before hand), scientific workflows are not necessarily the same as business workflows or personal pipelines, although they are similar. “Scientific workflows are widely recognised as a ‘useful paradigm to describe, manage, and share complex scientific analyses’,” says the Taverna website.

As you can already tell by the terminology, scientific workflows have become a separate entity, tied up in computational science. They range from simple shell scripts to heavy grid programs that enable distributed, parellel processing for large data sets. On the whole, the most useful workflows that are being used repeatedly by different labs and scientists are built within other programs designed for that purpose, which also means that they are portable, easily archived and accessed, and somewhat transparent as to their immediate function.
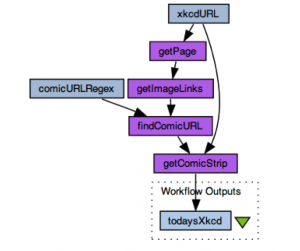
An example of a workflow workbench would be Taverna; others include Kepler, VisTrails, Pegasus, Knime, RapidMiner, and so on. These are programs which have been designed to make designing workflows easier, by using smaller components that can be added together, than just copying and pasting, or infitinitely fiddling with, long scripts of raw code. Here is an example workflow that gets a comic off of the popular xkcd site:
The workflows developed in these programs can be shared – and since 2007, there has been a single repository that has garned the most impact, partly due to the fact that the organisers also worked with or were the Kepler and Taverna developers, and partly because of APIs in those programs that access the site, and partly due to the lack of a single repository elsewhere (although there are others, such as Yahoo pipes, although that is not science based.) I am talking about myExperiment.org, which now boasts over 2000 uploaded workflows. These workflows have been uploaded entirely by the community, with no direct benefit to the uploader except to showcase work that might be helpful. As such, myExperiment is a wonderful textbook of open science at its best.
What did I do?
In my internship, I did several things РI mined google scholar, other websites concerning workflows, and various lists for pretty much all of the information I could get on workflows. All of these papers are now available in the Mendeley group I set up. The majority of these papers are by computational scientists Рgenerally bioinformatics, but also software developers Рabout workflows, with only a small subset being papers that have used workflows in their research and cite where they have uploaded them. As this is the case, and as the workflow systems I was working with had a very steep learning curve (somewhat unfortunately), it became apparent early on that a new approach was needed besides research.
So, I was tasked with getting all of the information I could off of myExperiment. They have an RDF backend that I found inscrutable, and that didn’t display the information very well once it was downloaded. To bypass this, I ended up screenscraping all of the information I could from the workflow pages themselves on myExperiment. This resulted in a fairly large amount of data on how scientists have been using myExperiment – which workflows were downloaded more, what the components were for those workflows, how they were tagged, what they did, and so on. This was the result of my internship, although the research is ongoing.
What did you discover?
Well, there were a lot of results. I ran hundreds of lines of R code (mostly because I was new to it and kept repeating myself.) I found that there are actually only a few people uploading tons of workflows onto myExperiment, which means that the main users of workflows may well be the developers. Most workflows aren’t very complex, at the end of the day, either – they do simple tasks. This has changed over time, but it’s hard to say whether that’s because of a change in the efficiency of the workbenches, or whether it’s because people are using simpler ones and tying them together (one of the important features of workflows is the ability to embed them inside each other). Most workflows are so-called ‘shims’, which means that they, effectively, change data from one format to another.
There were other results, of course, but I don’t want to give too much away as we’re trying to publish the results that we do have. Importantly, our resulting paper focuses around what we suggest for workflows based on our findings. Mainly, this settles down on a few core needs:
- community awareness – scientists dealing with large amounts of data or processing times need to be made aware that there are programs out there that can speed up their science, help them with feedback on their results, and make their efforts reproducible, which is one of the most important features of data-intensive science.
- standards – not just for workflow tags and names, but for workflow archiving and repository practices, for workflow usage, and most importantly for workflow referencing and citing in journals. If a paper mentions they use a workflow, it will go a long way towards helping reproducibility for other researchers, as well as understanding.
- education – there is a steep learning curve towards using workflows at the moment, and hopefully this could be passed by teaching students earlier about their use in helping with one’s own research, as well as to the wider community. What helps me do my work will help another successfully proof my work, which leads to better science, on the whole. It may also help others later do better work that will build off of mine. This is all good.
What’s next?
Well, I’m hoping to screen-scrape the groups, packs, tags, and user information off of myExperiment. I’m hoping to mine the research I have more and spot articles that reference workflows they use. I’m presenting my research in a couple of weeks at the DataONE All Hands Meeting in Albuquerque, NM, and I hope to get some feedback there to help with future studies. I am also hoping to run more R code on the information I have to figure out if there are clusters that show significantly that there are different sorts of workflows that are downloaded more often – say, bioinformatics ones as opposed to astronomical workflows (which do exist.) I hope that all of this will eventually end up being used to make the future of workflows brighter.
I am also hoping to setup a repository for workflows in Linguistics and social sciences, my own disciplines, as there is nothing of the sort at the moment, and very few people working on them. I am, to that end, currently trying to find a server to buy, and trying to set up an open access journal that would demand that the authors publish their code and datasets and not just their results. Hopefully, this will be a new and significant step in experimental social sciences towards better work. Also hopefully, some people will read this post and want to help out – if so, please let me know. I’m glad to say that this work has been a major learning experience, and that shows no sign of stopping.
Richard Littauer