OKFN at Open Science Summit 2011 #oss2011 – Day 1

Peter Murray Rust and I have just returned from representing OKFN at OSS 2011 in Mountain View. As a first time attendee, I was greatly impressed by the quality of the talks and ideas and the sheer enthusiasm in the room. You can check out Peter’s blog for a perspective from someone who has seen ideas presented last year blossom and real progress occur in many areas of open science.
As we’re all aware, open science is a broad church so I’ll try and pull out the highlights concerning open data/code and then list a few treats from the world of open scientific methods and DIY bio/hardware.

The conference kicked off with an excellent keynote from Victoria Stodden looking at transparency in the scientific method and the deep impact on dissemination of scientific knowledge. (Transcript; Slides [PDF]; Video). Victoria reminded us that transparency is nothing new in science, but the changing nature of scientific enquiry with the advent of computation and the data deluge leads to new issues of reproducibility. She described a credibility crisis for computational science due to a lack of open availability of code packages and appropriate descriptions of their use, meaning that many published results are neither verifiable nor verified. Reproducibility is one of the strongest arguments we have to encourage openness in science and the message was delivered very eloquently.
A take home message that set the Twitter feed alight was the idea that “an article (about computational science in Victoria’s talk – but applicable more generally) in a scientific publication is not the scholarship itself, it is merely advertising of the scholarship. The actual scholarship is the complete software development environment and the complete set of instructions which generated the figures.” Also read, complete methodology, datasets and instrumentation environment in other scientific disciplines.
Interestingly, in terms of how to encourage uptake of these ides and advocate open science, Victoria proposed sticking as closely as possible to scientific norms, which is undoubtedly sage advice but was slowly diverged from throughout the rest of the conference as more and more projects were presented that are reinventing the way science is performed and communicated, such as Matt Todd’s open source drug discovery project. Reproducibility and scientific integrity are familiar to scientists even if open science is not and these should form the core of our advocacy. The Panton Principles and the recently published Science Code Manifesto both emphasise these ideas strongly.
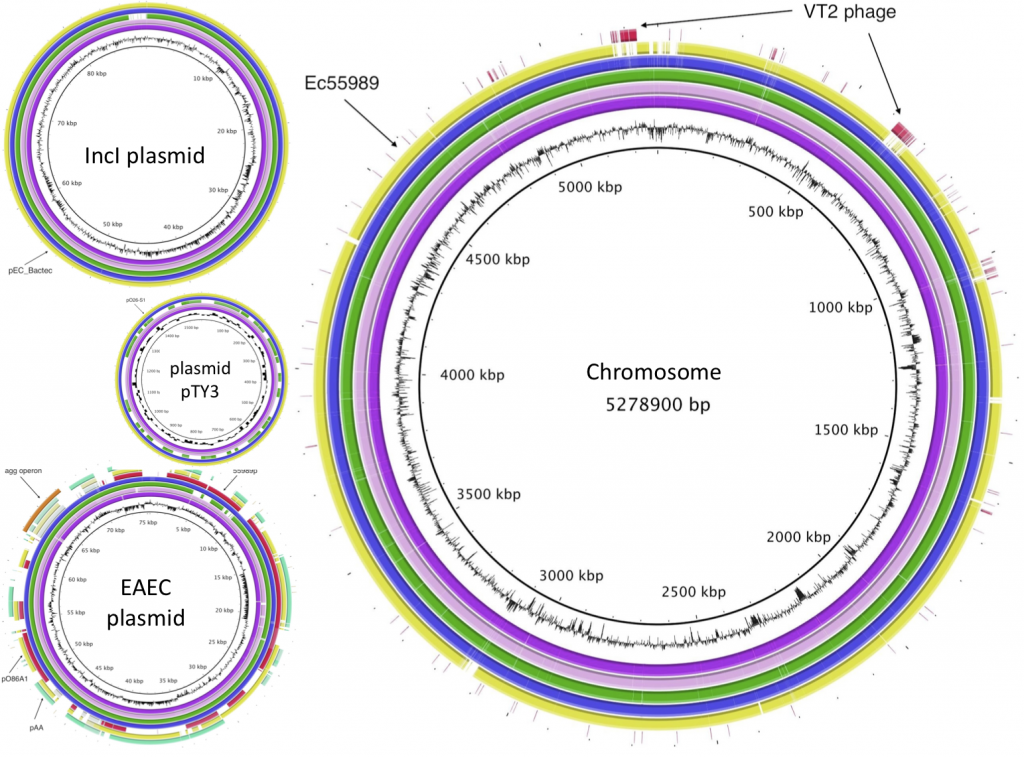
Edited highlights from open science in 2011 demonstrated how far we have come in the past year. A fascinating presentation from Joyce Peng of the Chinese genomics institute BGI (who have more DNA sequencing than the whole of the US government combined!) underplayed if anything their remarkable work on sequencing the highly pathogenic E.coli at the root of the European outbreak earlier this year. The raw sequence was completely openly shared on GitHub within 3 days of receiving a sample and high quality alignment and annnotation was completed in less than two weeks, with assistance and comments from hundreds of scientists around the globe who were able to freely access the data. However, subsequent publications (here and here) by two German groups failed to acknowledge this crowdsourcing effort.
Peter and I were up next and Peter led with the message that closed knowledge kills people – a situation that would likely have been the case in the E.coli outbreak where speed was of the essence. Indeed, Springer temporarily made 400 of its E.coli research papers freely available “to help researchers and medical professionals solve, or at the very least alleviate, this crisis”. Due to this speedy sharing and collaboration, the causative bacterial strain and the reason it was so pathogenic was quickly identified and openly published enabling contaminated bean sprouts to be identified as the carrier rather than the originally accused cucumber. More on our talk in a separate post, so I’ll move swiftly on to William Gunn from Mendeley, who pointed out the phenomenal growth in paper uploaded to their catalogue in the past year, with a total now standing at 120 million entries. Best of all, the bibliographic metadata associated with all of these articles is completely open (see the IsItOpenData? query!). Fore more about the importance and use of open bibliographic data see the Open Bibliography working group page.
OKFN’s very own Daniel Mietchen was up next, Skyping in to look at the integration of the wiki environment into every step of the research cycle in his role as Wikimedian in Residence on open science, based with OKFN Germany. You can browse through the wiki (naturally) for his talk and watch the video.
The final session of the day concerned open hardware and showed a fascinating glimpse into how 3D printing and bootstrapping low cost instruments can revolutionise the tools available to those outside academic institutions and big industry, such as small start ups and citizen scientists. The full video is available.
Rob Meagley makes £100 electrospray machines and $1000 plasma electorspray machines using a motley collection of glass sauce jars and other household objects. He is interested in nanomaterial structures.
The evening meetup was hosted by BioCurious, a biohack space in Mountain View where citizen scientists can make fluorescent GFP bacteria, sequence parts of their own genome and conduct other scientific experiments. As a molecular biologist I was impressed by the equipment they had in place, including three PCR machines.
A second hackspace we visited in San Francisco (Noisebridge) even had their own scanning electron microscope. The spirit of experimental discovery is alive and well and as Peter states on his blog “Science is for all of us, not just academia. Academia has recently made a very bad job of doing science on behalf of the community. Open Science shows that the vision is much larger.”
BGI E-coli image from http://bacpathgenomics.wordpress.com/2011/06/18/bgi-assembly-of-german-e-coli-outbreak-strain/

Leave a Reply