Second Quarterly Report on my Panton Fellowship
Peter Kraker - March 26, 2014 in Panton Fellowships
I am now almost halfway through my Panton Fellowship, so it is time to sum up my activities once again.
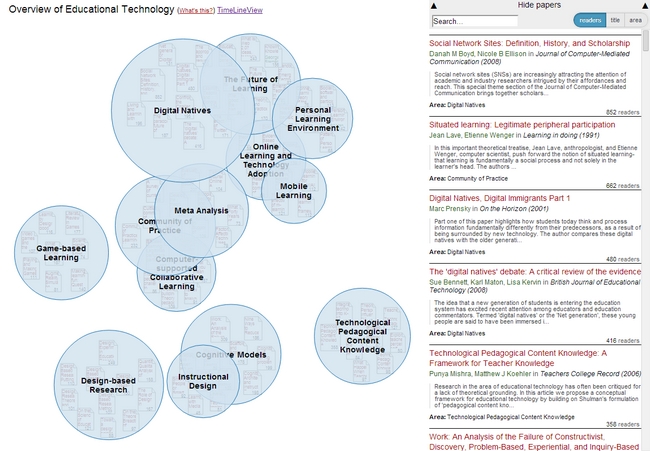
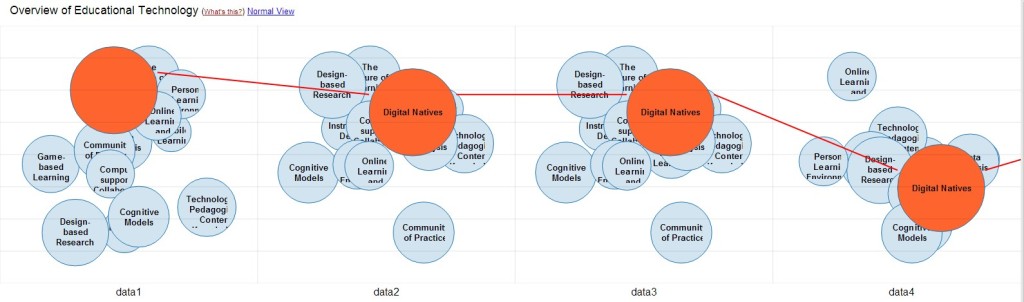
The most important activity in the last quarter was surely the work on the open source visualization Head Start. Head Start is intended to give scholars an overview of a research field. You can find out all about the initial release in this blog post. I was busy in the last few weeks with bugfixing and stability improvements. I also refactored the whole pre-processing system and further integrated the work of Philipp Weißensteiner with regards to time-series visualization. If you are interested in trying out Head Start, or – even better – would like to contribute to its development, check out the Github repository.
Furthermore, I attended the Science Online un-conference in Raleigh (February 27 to March 1). Scio14 was very inspiring and engaging. Cameron Neylon hosted a great session on imagining the far future of academic publishing. In Rachel Levy‘s workshop on visualizations, we reflected on our own visualizations and there were tons of tips for improving one’s work. Other great sessions included post-publication peer review (with Ivan Oransky), altmetrics (facilitated by Cesar Berrios-Otero), and alternate careers in science (led by Eva Amsen). I also encourage you to check out the videos of the keynotes which include a very inspiring talk by Rebecca Tripp and Meg Lowman on neglected audiences in science, and the awesone crowd-sourced 3D printing project for creating prosthetic hands by Nick Parker and Jon Schull.
Let’s move on to my work for the local Austrian community. Together with my fellow OKFN members Sylvia Petrovic-Majer, Stefan Kasberger, and Christopher Kittel, I became active (remotely for now) in the Open Access Network Austria (OANA). Specifically, I am contributing to the working group “Involvment of researchers in open access”. I am very excited about this opportunity as it is one of the objectives of my Panton Fellowship to draw more researchers in open science.
What else? Earlier this year, I was interviewed for the openscienceASAP podcast. In the interview, I talked about altmetrics, the need for an inclusive approach to open science, and the Panton Fellowships. You can find the podcast here (in German). If you have read my last report, you may remember that I spoke on a panel about open science at University of Graz. The video of the panel (in German) is now online and can be found here. Furthermore, I’d like to draw your attention to the monthly sum-ups of open science activities in the German speaking world and beyond: January, February.
So what will my next quarter look like? As you may remember from my last report, I am currently a visiting scholar at University of Pittsburgh. In the weeks to come, I will integrate Head Start with Conference Navigator 3, developed by the great folks of the PAWS Lab here in Pittsburgh. Conference Navigator is a nifty scheduling system that allows you to create a personal conference schedule by bookmarking talks from the program. The system then gives you recommendations for further talks based on your choices. Head Start will be used as an alternate way of looking at the topics of the conference, and to give better context to the talks that you already selected. I will return to Austria in June, just in time for Peter Murray-Rust‘s visit to Vienna. There are already a lot of activities planned around his stay, and I am very much looking forward to that. As always, please get in touch if you have any questions or comments, or in case you want to collaborate on one or the other project.