Open Training for Open Science
Jenny Molloy - December 21, 2014 in Featured, Reproducibility, Research, Tools
This is part of series of blog posts highlighting focus points for the Open Science Working Group in 2015. These draw on activities started in the community during 2014 and suggestions from the Working Group Advisory Board.
The Open Science Working group have long supported training for open science and early introduction of the principles of open and reproducible research in higher education (if not before!). This area was a focus in 2013-4 and grows in importance as we enter 2015 with the level of interest in openness in science increasing at a rapid rate. This post attempts to provide examples of training initiatives in which members of the working group have been involved and particular areas where work is lacking.
- Openness in higher education
- Strategies for training in open science
- The Open Science Training Initiative (OSTI)
- Developing Open Science Training Curricula
- Incorporating Open Science Training into Current Courses
- Conclusion
- Getting Involved in Open Science Training
Openness in higher education
Openness has the potential to radically alter the higher education experience. For instance, Joss Winn and Mike Neary posit that democratisation of participation and access could allow a reconstruction of the student experience in higher eduction to achieve this social relevance, they propose:
“To reconstruct the student as producer: undergraduate students working in collaboration with academics to create work of social importance that is full of academic content and value, while at the same time reinvigorating the university beyond the logic of market economics.”[1]
Openness focuses on sharing and collaboration for public good, at odds with the often competitive ethos in research and education. This involves more than simply implementing particular pedagogies or publishing open access articles – as Peters and Britez state bluntly in the first sentence of their book on open education:
“Open education involves a commitment to openness and is therefore inevitably a political and social project.” [2]
This could equally apply to open science. Openness is a cultural shift that is facilitated but not driven by legal and technical tools. In open education, for instance, open pedagogy makes use of now abundant openly licensed content but also places an emphasis on the social network of participants and the learner’s connections within this, emphasising that opening up the social institution of higher education is the true transformation. In open science, a lot of training focuses on the ability to manage and share research data, understand licensing and use new digital tools including training in coding and software engineering. However, understanding the social and cultural environment in which research takes place and how openness could impact that is arguably even more fundamental.
This section will focus on three topics around open science training, offering relevant linkages to educational literature and suggestions for teaching design:
- Use of open data and other open research objects in higher education.
- Use of open science approaches for research-based learning.
- Strategies for training in open science.
Strategies for training in open science
As openness is a culture and mindset, socio-cultural approach to learning and the construction of appropriate learning environments is essential. While the Winn and Neary [1] focus on the student as producer, Sophie Kay [3] argues that this can be detrimental as it neglects the role of students as research consumers which in turn neglects their ability to produce research outputs which are easily understood and reuseable.
Training in evolving methods of scholarly communication is imperative because there are major policy shifts towards a requirement for open research outputs at both the funder and learned society levels in the UK, EU and US. This is in addition to a growing grassroots movement in scientific communities, accelerated by the enormous shifts in research practice and wider culture brought about by pervasive use of the internet and digital technologies. The current generation of doctoral candidates are the first generation of `digital natives’, those who have grown up with the world wide web, where information is expected to be available on demand and ‘prosumers’ who consume media and information as well as producing their own via social media sites, are the norm. This norm is not reflected in most current scientific practice, where knowledge dissemination is still largely based on a journal system founded in the 1600s, albeit now in digital format. Current evidence suggests that students are not prepared for change, for example a major study of 17,000 UK graduate students [4] revealed that students:
- hold many misconceptions about open access publishing, copyright and intellectual property rights;
- are slow to utilise the latest technology and tools in their research work, despite being proficient in IT;
- influenced by the methods, practices and views of their immediate peers and colleagues.
While pre-doctoral training is just as important, the majority of open science training initiatives documented thus far have aimed at the early career research stage, including doctoral students.
The Open Science Training Initiative (OSTI)
Open Knowledge Panton Fellow Sophie Kay developed an Open Science Training Initiative (OSTI) [3], trialled in the Life Science Interface Doctoral Training Centre at the University of Oxford, which employs `rotation based learning’ (RBL) to cement the role of students as both producers and consumers of research through learning activities which promote the communication of coherent research stories that maximise reproducibility and usefulness. The content involves a series of mini-lectures around concepts, tools and skills required to practice openly, including an awareness of intellectual property rights and licensing, digital tools and services for collaboration, storage and dissemination, scholaraly communication and broader cultural contexts of open science.
The novel pedagogical approach employed was the creation of groups during an initiator phase where each group reproduces and documents a scientific paper, ensuring that outputs are in appropriate formats and properly licensed. Next the successor phase sees the reproduced work being rotated to another group who must again validate and build upon it in the manner of a novel research project, with daily short meetings with instructors to address any major issues. No intergroup communication is allowed during either phase, meaning that deficiencies in documentation and sticking points become obvious and hopefully leads to greater awareness among students of the adequacy of their future documentation. The pilot course involved 43 students and had a subject-specific focus on computational biology. Feedback was excellent with students feeling that they had learnt more about scientific working practises and indicating they were highly likely to incorporate ideas introduced during the course into their own practice.
This course design offers great scope for inter-institutional working and as it uses OERs the same training can be delivered in several locations but remains adaptable to local needs. RBL would be more challenging to mirror in wet labs but could be adapted for these settings and anyone is encouraged to remix and run their own instance. Sophie is especially keen to see the materials translated into further languages.
Developing Open Science Training Curricula
OSTI is one of the first courses to specifically address open science training but is likely the first of many as funding is becoming available from the European Commission and other organisations specifically aimed at developing open access and open science resources and pedagogies. Some of the key consideration for teaching design in this space are:
- How to address socio-cultural aspects in addition to imparting knowledge about legal and technical tools or subject-specific content and skills training.
- The current attitudes and perceptions of students towards intellectual property and the use of digital technologies and how this will impact their learning.
- The fast pace of change in policy requirements and researcher attitudes to aspects of open science.
- Additional time and resources required to run additional courses vs amelioration of existing activities.

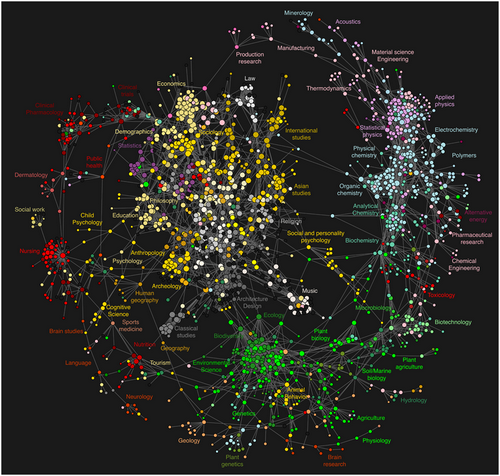
Open science curriculum map at MozFest 2014. Photo by Jenny Molloy, dedicated to the public domain via a CCZero waiver.
There are numerous one-off training events happening around the world, for instance the series of events funded by the FOSTER EU programme, which includes many workshops on open science. There are also informal trainings through organisations such as the Open Science working group local groups. Open science principles are incorporated into certain domain-specific conferences or skill-specific programmes like Software Carpentry Workshops, which have a solid focus on reproducibility and openness alongside teaching software engineering skills to researchers.
There are no established programmes and limited examples of open science principles incorporated into undergraduate or graduate curricula across an entire module or course. Historically, there have been experiments with Open Notebook Science, for instance Jean-Claude Bradley’s work used undergraduates to crowdsource solubility data for chemical compounds. Anna Croft from Bangor University presented her experiences encouraging chemistry undergraduates to use open notebooks at OKCon 2011 and found that competition between students was a barrier to uptake. At a graduate level, Brian Nosek has taught research methods courses incorporating principles of openness and reproducibility (Syllabus) and a course on improving research (Syllabus). The Centre for Open Science headed by Nosek also has a Collaborative Replications and Education Project (CREP) which is an excellent embodiment of the student as producer model and incorporates many aspects of open and reproducible science through encouraging students to replicate studies. More on this later!
It is clear that curricula, teaching resources and ideas would be useful to open science instructors and trainer at this stage. Billy Meinke and Fabiana Kubke helpfully delved into a skills-based curriculum in more depth during Mozilla Festival 2014 with their mapping session. Bill Mills of Mozilla Science Lab recently published a blog post on a similar theme and has started a pad to collate further information on current training programmes for open science. In the US, NCAES ran a workshop developing a curriculum for reproducible science followed by a workshop on Open Science for Synthesis .
NESCent ran a curriculum building workshop in Dec 2014 (see wiki). Several participants in the workshop have taught their own courses on Tools for Reproducible Research (Karl Broman) or reproducibility in statistics courses (Jenny Bryan). This workshop was heavily weighted to computational and statistical research and favoured R as the tool of choice. Interestingly their curriculum looked very different to the MozFest map, which goes to show the breadth of perspectives on open science within various communities of researchers!
All of these are excellent starts to the conversation and you should contribute where possible! There is a strong focus on data-rich, computational science so work remains to rethink training for the wet lab sciences. Of the branches of skills identified by Billy and Fabiana, only two of seven relate directly to computational skills, suggesting that there is plenty of work to be done! For further ideas and inspiration, the following section details some ways in which the skills can be further integrated into the curriculum through existing teaching activities.
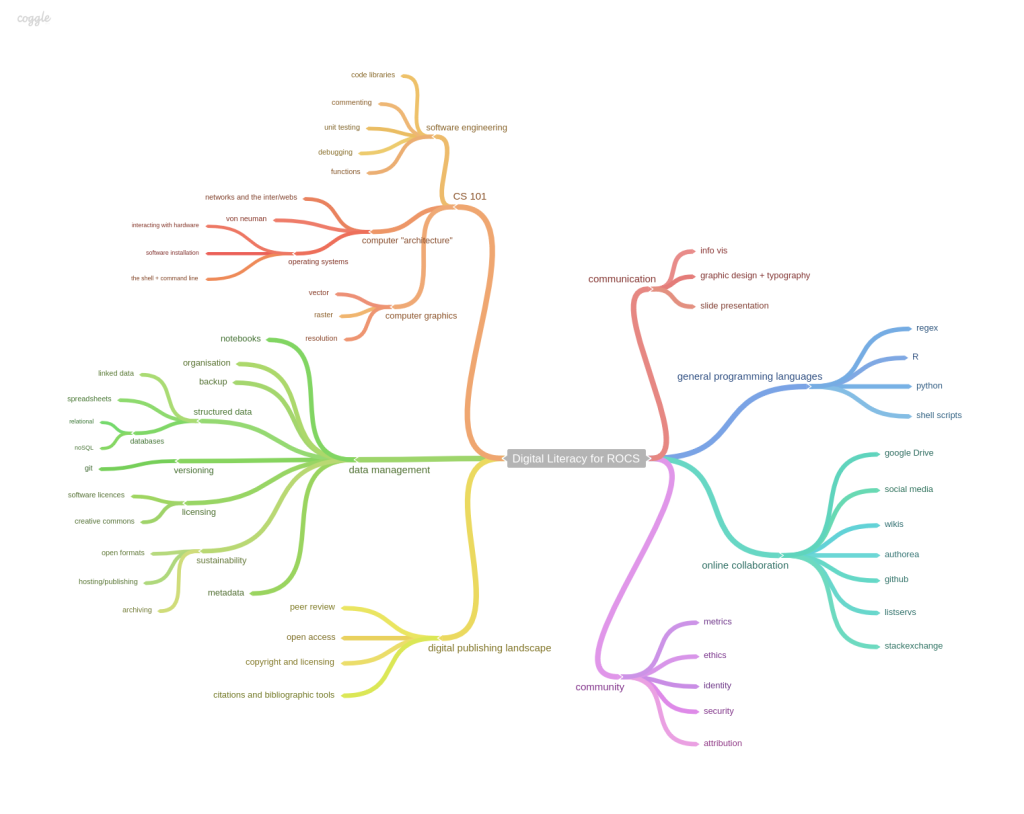
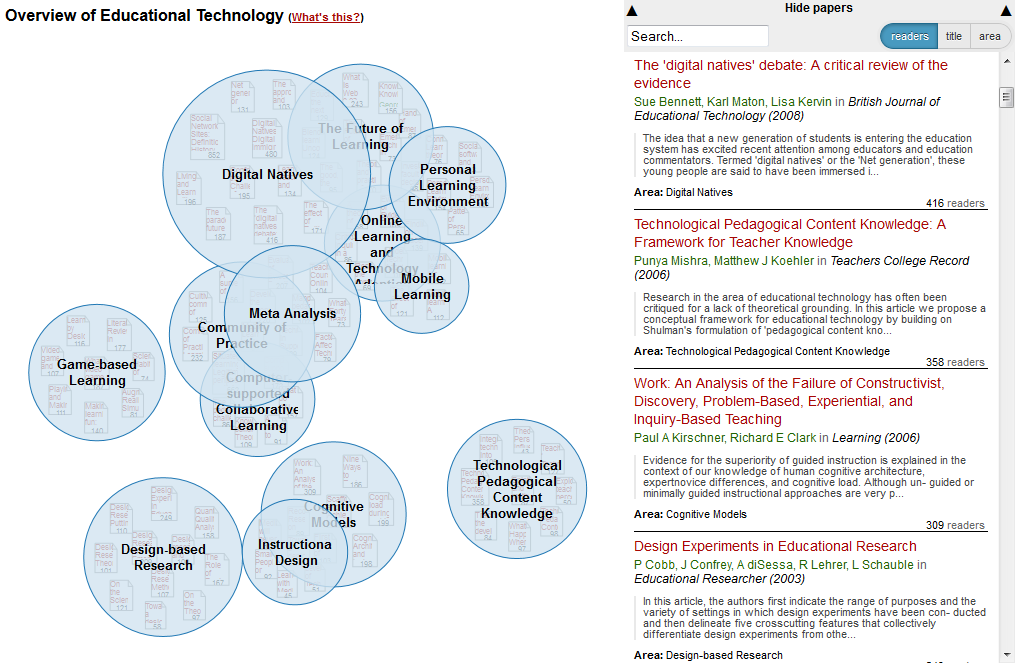
Skills map for Reproducible, Open and Collaborative Science. Billy Meinke and Fabiana Kubke’s session at MozFest 2014.
Incorporating Open Science Training into Current Courses
Using the open literature to teach about reproducibility
Data and software is increasingly published alongside papers, ostensibly enabling reproduction of research. When students try to reanalyse, replicate or reproduce research as a teaching activity they are developing and using skills in statistical analysis, programming and more in addition to gaining exposure to the primary literature. As much published science is not reproducible, limitations of research documentation and experimental design or analysis techniques may become more obvious, providing a useful experiential lesson.
There is public benefit to this type of analysis. Firstly, whether works are reproducible or not is increasingly of interest particularly to computational research and various standards and marks of reproducibility have been proposed but the literature is vast and there is no mechanism widely under consideration for systematic retrospective verification and demarcation of reproduciblity. Performing this using thousands of students in the relevant discipline could rapidly crowdsource the desired information while fitting easily into standard components of current curricula and offering a valid and useful learning experience.
The effect of `many eyes’ engaging in post-publication peer review and being trained in reviewing may also throw up substantive errors beyond a lack of information or technical barriers to reproduction. The most high profile example of this is the discovery by graduate student Thomas Herndon of serious flaws in a prominent economics paper when he tried to replicate its findings [5,6]. These included coding errors, selective exclusion of data and unconventional weighting of statistics, meaning that a result which was highly cited by advocates of economic austerity measures and had clear potential to influence fiscal policy was in fact spurious. This case study provides a fantastic example of the need for open data and the social and academic value of reanalysis by students, with the support of faculty.
This possibility has not been picked up in many disciplines but the aforementioned CREP project aims to perform just such a crowd-sourced analysis and asks instructors to consider what might be possible through student replication. Grahe et al., suggest that:
“Each year, thousands of undergraduate projects are completed as part of the educational experience…these projects could meet the needs of recent calls for increased replications of psychological studies while simultaneously benefiting the student researchers, their instructors, and the field in general.” [7]
Frank and Saxe [8] support this promise, reporting that they found teaching replication to be enjoyable for staff and students and an excellent vehicle for educating about the importance of reporting standards, and the value of openness. Both publications suggest approaches to achieving this in the classroom and are well worth reading for further consideration and discussion about the idea.
Reanalysing open data
One step from reproduction of the original results is the ability to play with data and code. Reanalysis using different models or varying parameters to shift the focus of the analysis can be very useful, with recognition of the limitations of experimental design and the aims of the original work. This leads us to the real potential for novel research using open datasets. Some fields lend themselves to this more than others. For example, more than 50% of public health masters projects across three courses examined by Feldman et al. [9] used secondary data for their analyses rather than acquiring expensive and often long-term primary datasets. Analysis of large and complex public health data is a vital graduate competency, therefore the opportunity to grapple with the issues and complexities of real data rather than a carefully selected or contrived training set is vital.
McAuley et al. [10] suggest that the potential to generate linked data e.g. interconnecting social data, health statistics and travel information, is the real power of open data and can produce highly engaging educational experiences. Moving beyond educational value, Feldman et al. [9] argue that open data use in higher education research projects allows for a more rapid translation of science to practise. However, this can only be true if that research is itself shared with the wider community of practise, as advocated by Lompardi [11]. This can be accomplished through the canonical scientific publishing track or using web tools and services such as the figshare or CKAN open data repositories, code sharing sites and wikis or blogs to share discoveries.
In order to use these digital tools that form the bedrock of many open science projects and are slowly becoming fully integrated into scholarly communication systems, technological skills and understanding of the process of knowledge production and disemmination in the sciences is required. Students should be able to contextualise these resources within the scientific process to prepare them for a future in a research culture that is being rapidly altered by digital technologies. All of these topics, including the specific tools mentioned above, are covered by the ROCS skills mapping from MozFest, demonstrating that the same requirements are coming up repeatedly and independently.
Use of open science approaches for research-based learning
There are several powerful arguments as to why engaging students in research-based activities leads to higher level and higher quality learning in higher education and the Boyer Commission on Educating Undergraduates in the Research University called for research based learning to become the standard, stating a desire:
“…to turn the prevailing undergraduate culture of receivers into a culture of inquirers, a culture in which faculty, graduate students, and undergraduates share an adventure of discovery.”
The previous section emphasised the potential role of open content, namely papers, data and code in research-based learning. In addition, the growing number of research projects open to participation by all – including those designated as citizen science – can offer opportunities to engage in research that scales and contributes more usefully to science than small research projects that may be undertaken in a typical institution as part of an undergraduate course. These open science activities offer options for both wet and dry lab based activities in place or in addition to standard practical labs and field courses.
The idea of collaborative projects between institutions and even globally is not new, involvement in FOSS projects for computational subjects has long been recognised as an excellent opportunity to get experience of collaborative coding in large projects with real, often complex code bases and a `world-size laboratory’ [12]. In wet lab research there are examples of collaborative lab projects between institutions which have been found to cut costs and resources as well as increasing the sample size of experiments performed to give publishable data [13]. Openness offers scaling opportunities to inter-institutional projects which might otherwise not exist by increasing their visibility and removing barriers to further collaborative partners joining.
There are several open and citizen science projects which may offer particular scope for research-based learning. One could be the use of ecology field trips and practicals to contribute to the surveys conducted by organisations such as the UK Biological Records Centre, thus providing useful data contributions and access to a wider but directly relevant dataset for students to analyse. NutNet is a global research cooperative which sets up node sites to collect ecosystem dynamics data using standard protocols for comparison across sites globally, as this is a longitudinal study with most measurements being taken only a couple of times a year it offers good scope for practical labs. On a more ad hoc basis, projects such as Open Source Malaria offer many project and contribution opportunities e.g. a request to help make molecules on their wishlist and a GitHub hosted to do list. One way of incorporating these into curricula are team challenges in a similar vein to the iGEM synthetic biology project, which involves teams of undergraduates making bacteria with novel capabilities and contributes the DNA modules engineered to a public database of parts known as BioBricks.
In conclusion, open and citizen science projects which utilise the internet to bring together networks of people to contribute to live projects could be incorporated into inquiry-based learning in higher education to the benefit of both students and the chosen projects, allowing students to contribute truly scientifically and socially important data in the `student as producer’ model while maintaining the documented benefits of research-based pedagogies. This ranges from controlled contributions to practice particular skills through discovery-oriented tasks and challenges such as iGEM, allowing students to generate research questions independently.
There are significant challenges in implementing these types of research-based activities, many of which are true of `non-open’ projects. For instance, there are considerations around mechanisms of participation and sharing processes and outputs. Assessment becomes more challenging as students are collaborating rather than providing individual evidence of attainment. As work is done in the open, provenance and sharing of ideas requires tracking.
Conclusion
This post has introduced some ideas for teaching open science focusing on the student as both a producer and consumer of knowledge. The majority of suggestions have centred around inquiry-based learning as this brings students closer to research practices and allows social and cultural aspects of science and research to be embedded in learning experiences.
Explicitly articulating the learning aims and values that are driving the teaching design would be useful to enable students to critique them and arrive at their own conclusions about whether they agree with openness as a default condition. There is currently little systematic evidence for the proposed benefits of open science, partly because it is not widely practised in many disciplines and also as a result of the difficulty of designing research to show direct causality. Therefore, using evidence-based teaching practices that attempt to train students as scientists and critical thinkers without exposing the underlying principles of why and how they’re being taught would not be in the spirit of the exercise.
Support for increased openness and a belief that it will lead to better science is growing, so the response of the next generation of scientists and their decision about whether to incorporate these practices into their work has great implications for the future research cultures and communities. At the very least, exposure to these ideas during under- and postgraduate training will enable students to be aware of them during their research careers and make more informed decisions about their practises, values and aims as a researcher. There are exciting times ahead in science teaching!
If you’ve found this interesting, please get involved with a growing number of like-minded people via the pointers below!
Getting Involved in Open Science Training
- Run an OSTI in your institution. Contact Sophie Kay for more info via the OSTI website.
- Contribute to the Mozilla Science pad on an open science curriculum. All are welcome!
- Contribute to the Centre for Open Science Collaborative Replications and Education Project (CREP).
More projects people could get involved with? Add them to the comments and the post will be updated.
References
- Neary, M., & Winn, J. (2009). The student as producer: reinventing the student experience in higher education.
- Peters, M. A., & Britez, R. G. (Eds.). (2008). Open education and education for openness. Sense Publishers.
- For a peer-reviewed paper on the OSTI initiative, see Kershaw, S.K. (2013). Hybridised Open Educational Resources and Rotation Based Learning. Open Education 2030. JRC−IPTS Vision Papers. Part III: Higher Education (pp. 140-144). Link to the paper in Academia.edu
- Carpenter, J., Wetheridge, L., Smith, N., Goodman, M., & Struijvé, O. (2010). Researchers of Tomorrow: A Three Year (BL/JISC) Study Tracking the Research Behaviour of’generation Y’Doctoral Students: Annual Report 2009-2010. Education for Change.
- Herndon, T., Ash, M., & Pollin, R. (2014). Does high public debt consistently stifle economic growth? A critique of Reinhart and Rogoff. Cambridge journal of economics, 38(2), 257-279.
- Roose, Kevin. (2013). Meet the 28-Year-Old Grad Student Who Just Shook the Global Austerity Movement}. New York Magazine. Available from http://nymag.com/daily/intelligencer/2013/04/grad-student-who-shook-global-austerity-movement.html. Accessed 20 Dec 2014.
- Grahe, J. E., Reifman, A., Hermann, A. D., Walker, M., Oleson, K. C., Nario-Redmond, M., & Wiebe, R. P. (2012). Harnessing the undiscovered resource of student research projects. Perspectives on Psychological Science, 7(6), 605-607.
- Frank, M. C., & Saxe, R. (2012). Teaching replication. Perspectives on Psychological Science, 7(6), 600-604.
- Feldman, L., Patel, D., Ortmann, L., Robinson, K., & Popovic, T. (2012). Educating for the future: another important benefit of data sharing. The Lancet, 379(9829), 1877-1878.
- McAuley, D., Rahemtulla, H., Goulding, J., & Souch, C. (2012). 3.3 How Open Data, data literacy and Linked Data will revolutionise higher education.
- Lombardi, M. M. (2007). Approaches that work: How authentic learning is transforming higher education. EDUCAUSE Learning Initiative (ELI) Paper, 5.
- O’Hara, K. J., & Kay, J. S. (2003). Open source software and computer science education. Journal of Computing Sciences in Colleges, 18(3), 1-7.
- Yates, J. R., Curtis, N., & Ramus, S. J. (2006). Collaborative research in teaching: collaboration between laboratory courses at neighboring institutions. Journal of Undergraduate Neuroscience Education, 5(1), A14.
Licensing
Text is licensed under the Creative Commons CC0 1.0 Universal waiver. To the extent possible under law, the author(s) have dedicated all copyright and related and neighbouring rights to this text to the public domain worldwide.
Open Books Image: by opensourceway on Flickr under CC-BY-SA 2.0








 Tyng-Ruey Chuang tackled some of these issues in his talk on ‘
Tyng-Ruey Chuang tackled some of these issues in his talk on ‘